splitcode
splitcode is a tool for flexible, efficient parsing, interpreting, and editing of technical sequences in sequencing reads.
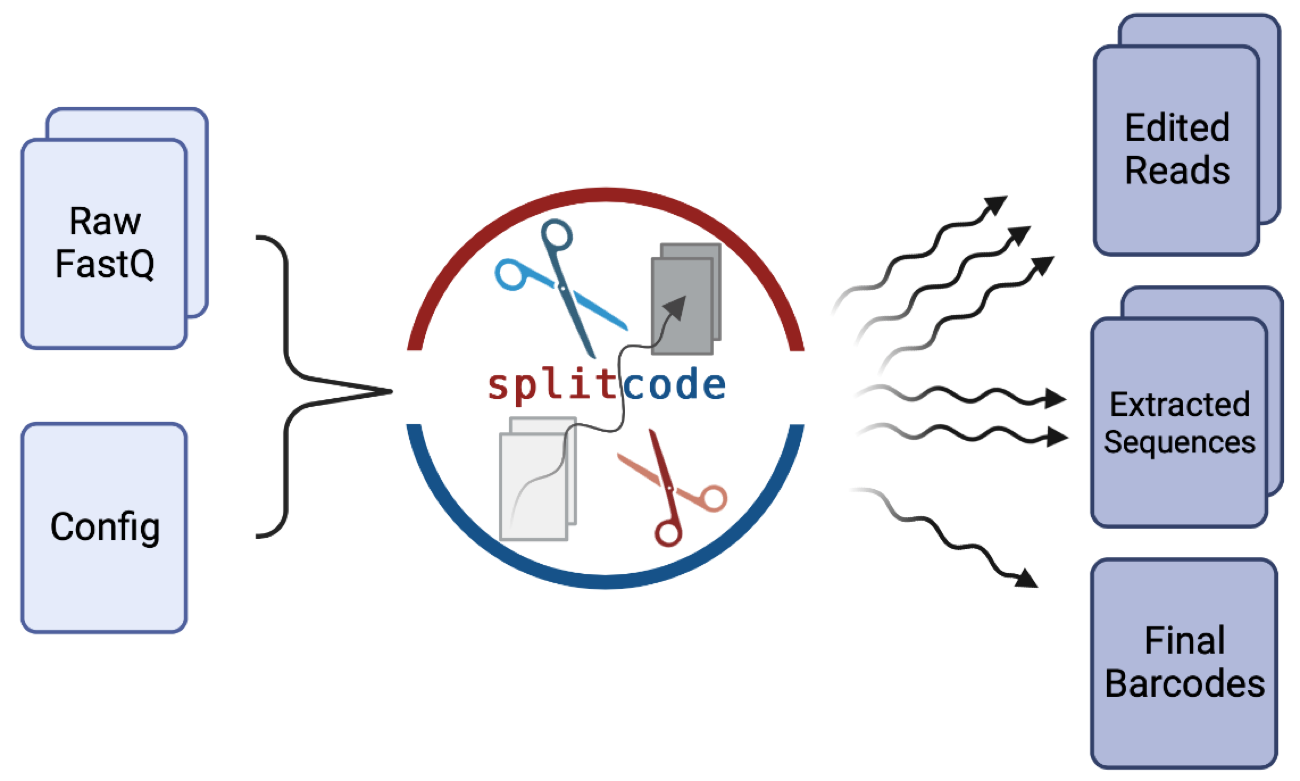
Introduction:
Contents:
Tutorials:
Citation:
Sullivan DK, Pachter L. (2023). Flexible parsing and preprocessing of technical sequences with splitcode. bioRxiv 2023.03.20.533521. https://doi.org/10.1101/2023.03.20.533521